Introduction
Artificial intelligence is changing the way we analyze complex biological data, and deep neural networks (DNNs) are at the heart of this technology. But what exactly is a DNN, and how can we use it to process something like T-cell receptor (TCR) sequences?
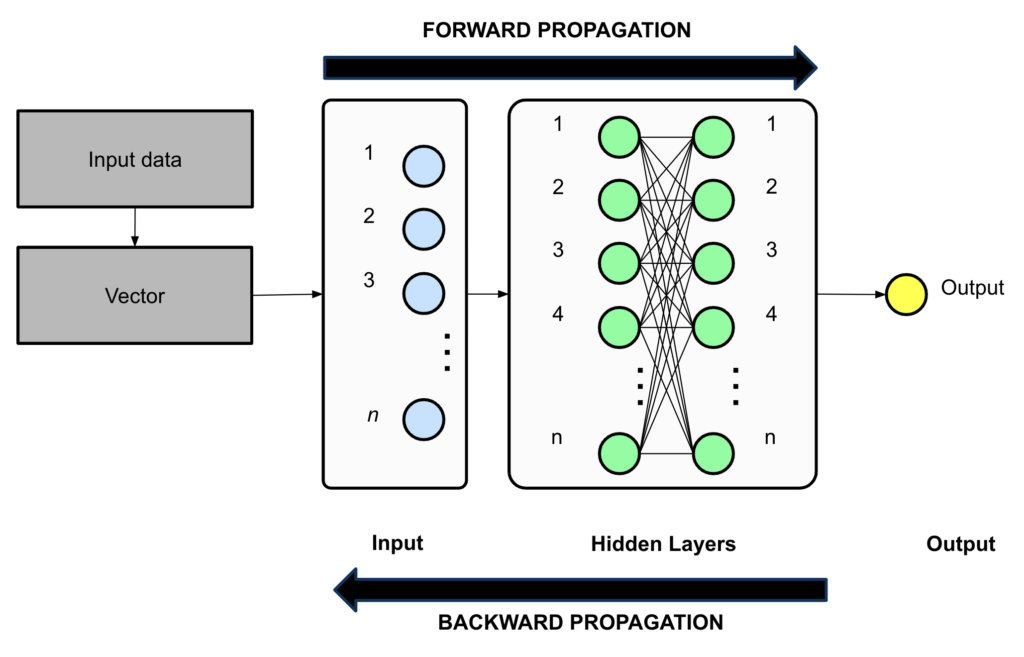
A deep neural network is a computational model consisting of layers of interconnected nodes. Each node applies mathematical operations to incoming data, transforming it as it moves through the network.
The key components of a deep neural network are:
- The Input Layer – where data enters the network
- Hidden Layers – where computations happen
- Output Layer – where predictions or classifications are made
From Amino Acids to Vectors
Neural networks process numerical data, but TCR sequences are strings of letters representing amino acids. How do we convert these sequences into a format that a neural network can understand?
One common approach is the one-shot representation (Step 1 in Figure 1). Each unique amino acid in the sequence is represented as a vector of 0s and 1s. For example, the amino acid sequence A-C-G-T-Y could be encoded as the 5 X 5 matrix:
A [1, 0, 0, 0, 0]
C [0, 1, 0, 0, 0]
G [0, 0, 1, 0, 0]
T [0, 0, 0, 1, 0]
Y [0, 0, 0, 0, 1]
A full TCR sequence is then represented as a series of these vectors, forming an input data matrix.
Forward Propagation: How Data Moves Through the Network
Once we have our input vector representation, we feed it into the first layer of the neural network. This data moves forward through each hidden layer in a process called forward propagation (Step 2 in Figure 1).
| In each node of a hidden layer | Zi = weight * [x1, x2, x3, x4…..xn] + bias |
| Weight and bias are adjustable parameters | |
| Zi is the linear manipulation of the one-shot vector |
At each layer, the data is multiplied by learned weights, and biases are applied to adjust the data for the next layer. These transformations allow the network to extract relevant features from the data.
The result of this calculation is passed to an activation function, which introduces non-linearity to the network’s output. Without an activation function, neural networks would simply perform linear transformations, limiting their ability to model complex patterns.
The Activation Function
Without an activation function, neural networks would just be linear regressions. But real world data- whether its images, text, or biological sequences- is highly non-linear.
| Activation Function | Acts on Zi in each node of a hidden layer. This introduces non-linearity, example of the sigmoid function: |
| a(Zi) = (1 + e-z) -1 |
The activation function is key for the network to learn complex patterns and relationships in the data.
The Loss Function Facilitates Learning
After passing through several hidden layers, the network produces an output (Step 3 in Figure 1)—for example: a classification of TCR sequences based on their immune function. But how does the network know if its prediction is correct?
This is where the loss function comes in. The loss function measures the difference between the predicted output and the true label from the training data. Common loss functions include cross-entropy loss for classification problems.
The goal of training is to minimize this loss by adjusting the weights and biases in the network, which is done through backpropagation (Step 4 in Figure 1). Backpropagation calculates gradients to determine how to update each parameter to reduce the error.
Conclusion
Deep neural networks provide a powerful way to analyze complex biological data, such as TCR sequences. By converting sequences into numerical vectors, applying weighted transformations, passing data through activation functions, and optimizing with loss functions, DNNs can uncover patterns that are beyond human intuition. However, DNNs require large datasets to train effectively and can be computationally expensive, making them most useful in environments with substantial data.
Outsourcing Bioinformatics Analysis: How Bridge Informatics (BI) Can Help
We are passionate about empowering life science companies with cutting-edge technologies. BI’s data scientists prioritize studying, understanding, and reporting on the latest developments so we can advise our clients confidently. Our bioinformaticians are trained bench biologists, so they understand the biological questions driving your computational analysis.
From pipeline development and software engineering to assembling Deep Neural Networks, and deploying such bioinformatic tools, BI can help you on every step of your research journey. As experts across data types from leading sequencing platforms, we can help you tackle the challenging computational tasks of storing, analyzing and interpreting genomic and transcriptomic data. Click here to schedule a free introductory call with a member of our team.
